Computational Analysis of Social Complexity
Fall 2025, Spencer Lyon
Prerequisites
- Julia setup
- Julia basics
- Julia types and methods
Outcomes
- Understand key components of networks/graphs
- Use the Graphs.jl package for working with graphs in Julia
- Implement the breadth-first search algorithm
References
- Easley and Kleinberg chapter 2
Introduction¶
Why Study Graphs?¶
- Economic, cultural, political, and social interactions are influenced by structure of relationships
- Transmission of viruses
- International trade, supply chains, marketplaces
- Spread of information, diffusion of innovation
- Political persuasion, localized voting patterns
- Human behaviors influenced by network of friends (sports, clothes, music)
- Behaviors can be effected by social networks
- “Influencers”
- Circles of followers can create echo chambers
Edges and Nodes¶
- A graph specifies relationships between a collection of items
- Each item is called a node
- A relationship between nodes is represented by an edge
- Visually, graphs might look like this:
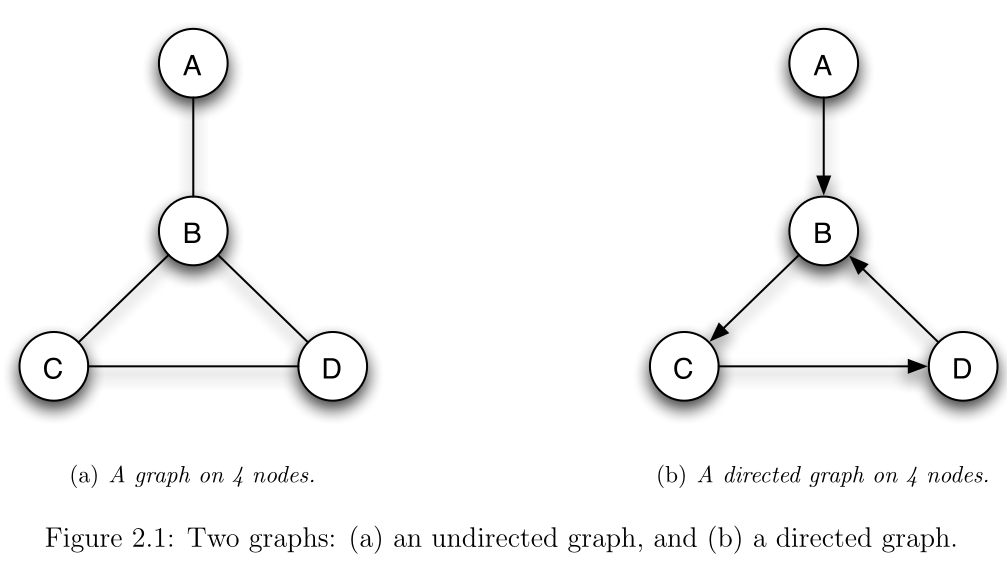
- Here the nodes are
A,B,C,D - The edges connect nodes
A-B,B-C,B-D,C-D
Adjacency Matrix¶
- How might we represent the graph above numerically?
- One very common approach is to use a matrix of 0’s and 1’s called an adjancency matrix
- Suppose we have a graph of nodes
- Without loss of generality, we’ll represent them as integers
1:N
- Without loss of generality, we’ll represent them as integers
- Let be our adjacency matrix
- Element will be zero unless there is an edge between nodes and (diagonal is left as 0)
- In our above we had
- Nodes
A,B,C,D(or 1, 2, 3, 4 respectively) - Edges connecting nodes
1-2,2-3,2-4,3-4
- Nodes
- The adjacency matrix for this example is
Graphs in Julia¶
- In Julia there are a few ways we could represent our example graph above
- We could start with the adjacency matrix concept as follows
A = [
0 1 0 0
1 0 1 1
0 1 0 1
0 1 1 0
]4×4 Matrix{Int64}:
0 1 0 0
1 0 1 1
0 1 0 1
0 1 1 0Working with Adjacency Matrices¶
- An adjacency matrix gives us a lot of information about the structure of the graph
- We could compute all of the following
- Total number of nodes: number of rows or columns of
- Total number of edges:
- Node with most edges:
- Average number of edges per node:
Exercise: Adjacency Matrix¶
- In the cell below we have defined an adjacency matrix called
A_ex1 - Using
A_ex1answer the following questions:- How many nodes are in the graph?
- How many edges?
- Node with most edges (hint, use the
dimsargument tosumand then theargmaxfunction) - Average number of edges per node
- Number of connections for node 7:
import Random
Random.seed!(42)
A_ex1 = zeros(Int, 30, 30)
for i in 1:30
for j in (i+1):30
if rand() < 0.2
A_ex1[i, j] = 1
A_ex1[j, i] = 1
end
end
end# Your code here
ex1_total_nodes = missing
ex1_total_edges = missing
ex1_node_most_edges = missing
ex1_average_edges_per_node = missing
ex1_connections_node_7 = missingmissingGraphs.jl¶
- There are many smart graph theory experts in the Juila community
- They have built a package called Graphs for working with graphs (as well as ancillary pacakges for extra features)
- We can build a
Graphs.Graphobject directly from our adjacency matrix
using GraphsG1 = Graph(A){4, 4} undirected simple Int64 graphcollect(edges(G1)) # collect turns an `iterator` into an array4-element Vector{Graphs.SimpleGraphs.SimpleEdge{Int64}}:
Edge 1 => 2
Edge 2 => 3
Edge 2 => 4
Edge 3 => 4collect(vertices(G1)) # Graphs refers to nodes as `vertices`4-element Vector{Int64}:
1
2
3
4Visualizing Graphs¶
- We can use the GraphPlot package to visualize our graph
- Note that the actual placement of the nodes is randomly generated and then tweaked to clearly show all nodes and edges
- The important thing is not the placement of nodes, but rather their relative structure
using GraphPlot # load GraphPlot packagegplot(G1)Loading...
Size considerations¶
- Using
Array{Int64,2}to store an adjacency matrix turns out to be a rather costly way to store a graph - In the original example graph we had 4 nodes and 4 edges
- To store this we needed to have a 4x4 matrix of 64 bit integers
- This is only (
Int(16 * 64 / 8) == 128) bytes in our exapmle, - But consider a graph of websites and links between them -- that graph would have millions of nodes and edges...
- This is only (
- There are a few approaches to reducing this storage cost:
- Only store the upper triangle of the matrix
- Use
Array{Bool,2}instead ofArray{Int64,2}to store adjacency matrix ( each element onlysizeof(Bool) == 1bit!) - Use a SparseMatrix
- Store as a
Vector{Vector{Int}}
# Vector{Vector{Int}}
A2 = [[2], [1, 3, 4], [2, 4], [2, 3]]4-element Vector{Vector{Int64}}:
[2]
[1, 3, 4]
[2, 4]
[2, 3]G1.fadjlist4-element Vector{Vector{Int64}}:
[2]
[1, 3, 4]
[2, 4]
[2, 3]Graph Theory Concepts¶
- Let’s explore some concepts often used in analysis of graphs
Paths¶
- When studying graphs it is often natural to ask about how things travel or flow across the graph
- For example, how information spreads amongst a group of friends, how data travels the internet, how diseases are transmitted from one person to another, and how people navigate a metro subway system
- In each of these cases, the flow of things goes from node to node across edges
- A flow from one any node to another node is called a path
Degree¶
- The degree of a node is the number of edges connected to it
- In our example graph G1, node B has degree 3 (connected to A, C, and D)
- Node A has degree 1 (only connected to B)
- Degree is a fundamental measure of node importance in a network
- High-degree nodes are often called “hubs”
- In social networks, these might be influential people or “connectors”
- We can compute degree directly from the adjacency matrix
- Degree of node is
- Graphs.jl provides the
degreefunction
degree(G1)4-element Vector{Int64}:
1
3
2
2degree(arpa, node_ints["MIT"])UndefVarError: `arpa` not defined in `Main`
Suggestion: check for spelling errors or missing imports.
Stacktrace:
[1] top-level scope
@ ~/Teaching/UCF/CAP-6318/book-myst/week03/jl_notebook_cell_df34fa98e69747e1a8f8a730347b8e2f_Y164sZmlsZQ==.jl:1- The degree distribution of a network tells us a lot about its structure
- Many real-world networks have a few high-degree hubs and many low-degree nodes
- Social networks: celebrities with millions of followers
- Internet: major websites with many incoming links
- Transportation: hub airports like Atlanta or Chicago
Arpanet Example¶
- Consider the following Graph of the first iteration of the internet
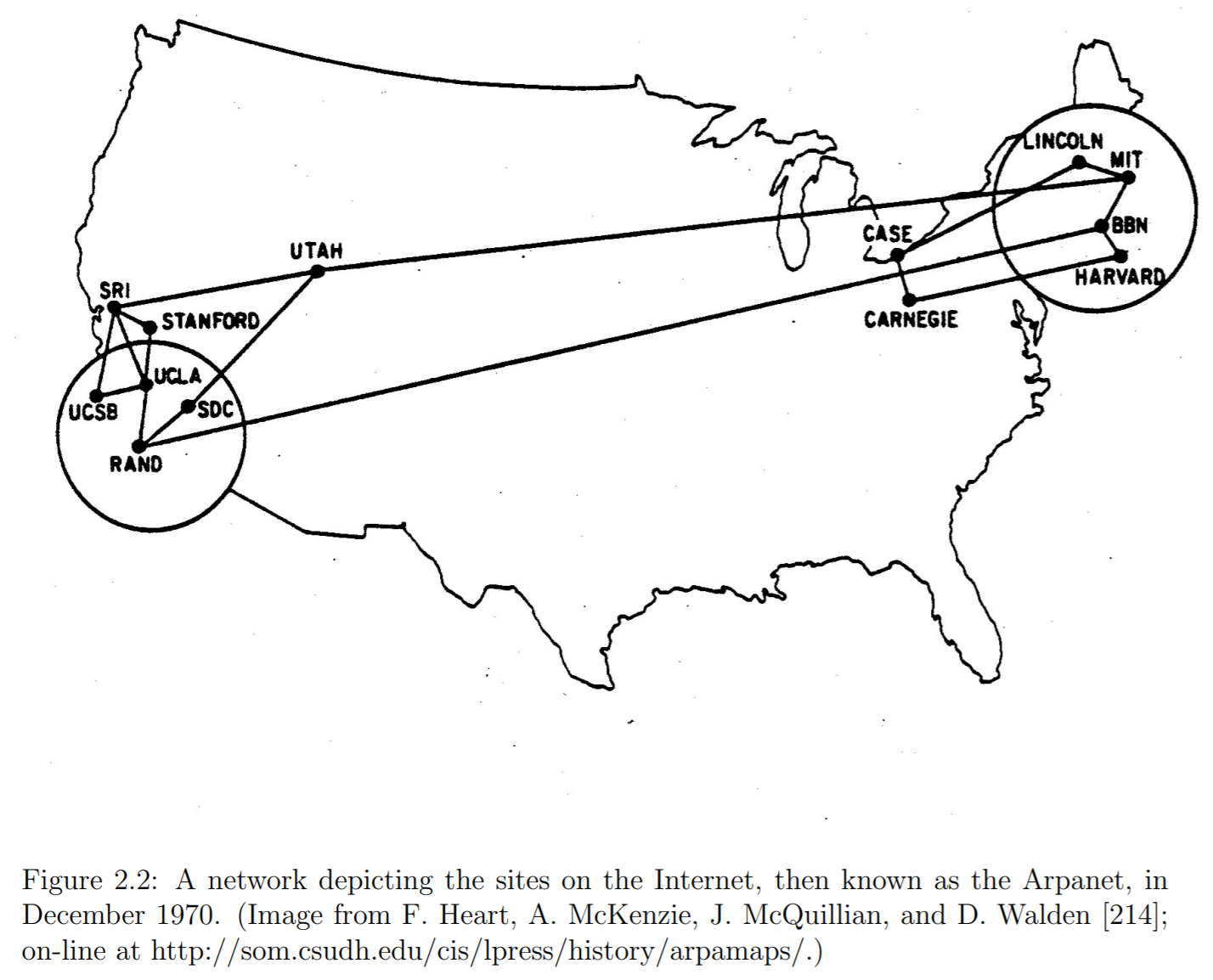
- There are many possible paths through this network
- Consider a path from UCSB to MIT:
UCSB-UCLA-RAND-BBN-MIT - Another possible path from UCSB to MIT is
UCSB-SRI-UTAH-MIT
Graphs.jl Arpanet¶
- Let’s define the Arpanet using Graphs as it will be helpful throughout this lecture
nodes = [
"UCSB" => ["SRI", "UCLA"],
"SRI" => ["UCSB", "UCLA", "STAN", "UTAH"],
"UCLA" => ["SRI", "UCSB", "STAN", "RAND"],
"STAN" => ["SRI", "UCLA"],
"UTAH" => ["SRI", "SDC", "MIT"],
"SDC" => ["UTAH", "RAND"],
"RAND" => ["UCLA", "SDC", "BBN"],
"MIT" => ["UTAH", "BBN", "LINC"],
"BBN" => ["MIT", "RAND", "HARV"],
"LINC" => ["MIT", "CASE"],
"CASE" => ["LINC", "CARN"],
"CARN" => ["CASE", "HARV"],
"HARV" => ["CARN", "BBN"]
]
node_ints = Dict(zip(first.(nodes), 1:length(nodes)))
arpa = SimpleGraph(length(nodes))
for (node, edges) in nodes
for e in edges
add_edge!(arpa, node_ints[node], node_ints[e])
end
end
# save graph for loading in future
savegraph("arpanet.lg", arpa)1arpa{13, 17} undirected simple Int64 graphgplot(arpa, nodelabel=first.(nodes))Loading...
Cycles¶
- An important concept when analyzing graphs is the concept of a cycle
- A cycle is a path that starts and ends at the same node
- For the ARPA net, an example cycle is
LINC-CASE-CARN-HARV-BBN-MIT-LINC - Question... what is the shortest possible cycle in a graph (including all endpoints)?
- Cycles appear frequently in real-world networks
- Friendship networks: if A is friends with B, and B with C, often A and C become friends
- Transportation networks: cyclical routes allow for redundancy and flexibility
- Supply chains: cyclical dependencies can create both resilience and vulnerabilities
Trees¶
- A tree is a connected graph with no cycles
- Trees have exactly N-1 edges for N nodes (the minimum to stay connected)
- Adding any edge to a tree creates a cycle
- Removing any edge from a tree disconnects it
- Trees appear frequently in hierarchical structures
- Organizational charts: CEO at root, departments as branches
- Family trees: ancestors and descendants
- Decision trees: choices branching from initial state
- File systems: directories and subdirectories
- The breadth-first search we’ll see later naturally creates a tree from any graph
# Example: create a simple tree
tree = SimpleGraph(5)
add_edge!(tree, 1, 2)
add_edge!(tree, 1, 3)
add_edge!(tree, 2, 4)
add_edge!(tree, 2, 5)
gplot(tree, nodelabel=1:5)Loading...
is_tree(tree) # Graphs.jl can check if a graph is a treetrueis_tree(G1) # Our original graph has a cycle, so it's not a treefalseConnectedness¶
- A graph is connected if there exists a path between every pair of nodes
- In other words, you can travel from any node to any other node by following edges
- The ARPA network was designed to be connected -- ensuring communication between all sites
- We can check if a graph is connected using Graphs.jl
is_connected(arpa)true- It is natural to believe that many real-world networks are connected
- Transportation: you can get to any station
- Internet: you can visit any website
- But it is entirely possible to have a non-connected graph
- Social networks (nodes: people, edges: friendships) of college students who different countries
- Suppliers for a textile company vs a microchip manufacturer
Distance¶
- We can extend concept of paths between nodes, to include a notion of distance
- The length of a path is the number of steps it takes from beginning to end
MIT-BBN-RAND-UCLAhas length 3 (starting fromMITtake three steps before ending atUCLA)
- The distance between two nodes, is the length of the shortest path between those nodes
- Graphs can compute distances using the
gdistancesfunction - Below we compute the distance between
UCLAand all nodes
Dict(zip(first.(nodes), gdistances(arpa, node_ints["UCLA"])))Dict{String, Int64} with 13 entries:
"HARV" => 3
"UTAH" => 2
"UCSB" => 1
"LINC" => 4
"RAND" => 1
"MIT" => 3
"CASE" => 5
"SRI" => 1
"STAN" => 1
"UCLA" => 0
"SDC" => 2
"CARN" => 4
"BBN" => 2Breadth-First Search¶
- If asked, how would you go about computing the distance between the
HARVnode and all other nodes? - One iterative approach might be:
- Start with
HARV: note it is distance zero toHARV - Move on to all nodes directly connected to
HARV: these are distance 1 - Then move to all nodes connected to nodes that are distance 1 from
HARV(excluding any you may have already found): declare these to be at distance 2 fromHARV - Continue traversing edges until you have visited all nodes
- Start with
- This algorihtm is called breadth-first search
Example: Breadth-First Search from MIT¶
- The image below shows how breadth-first search would proceed for the MIT node
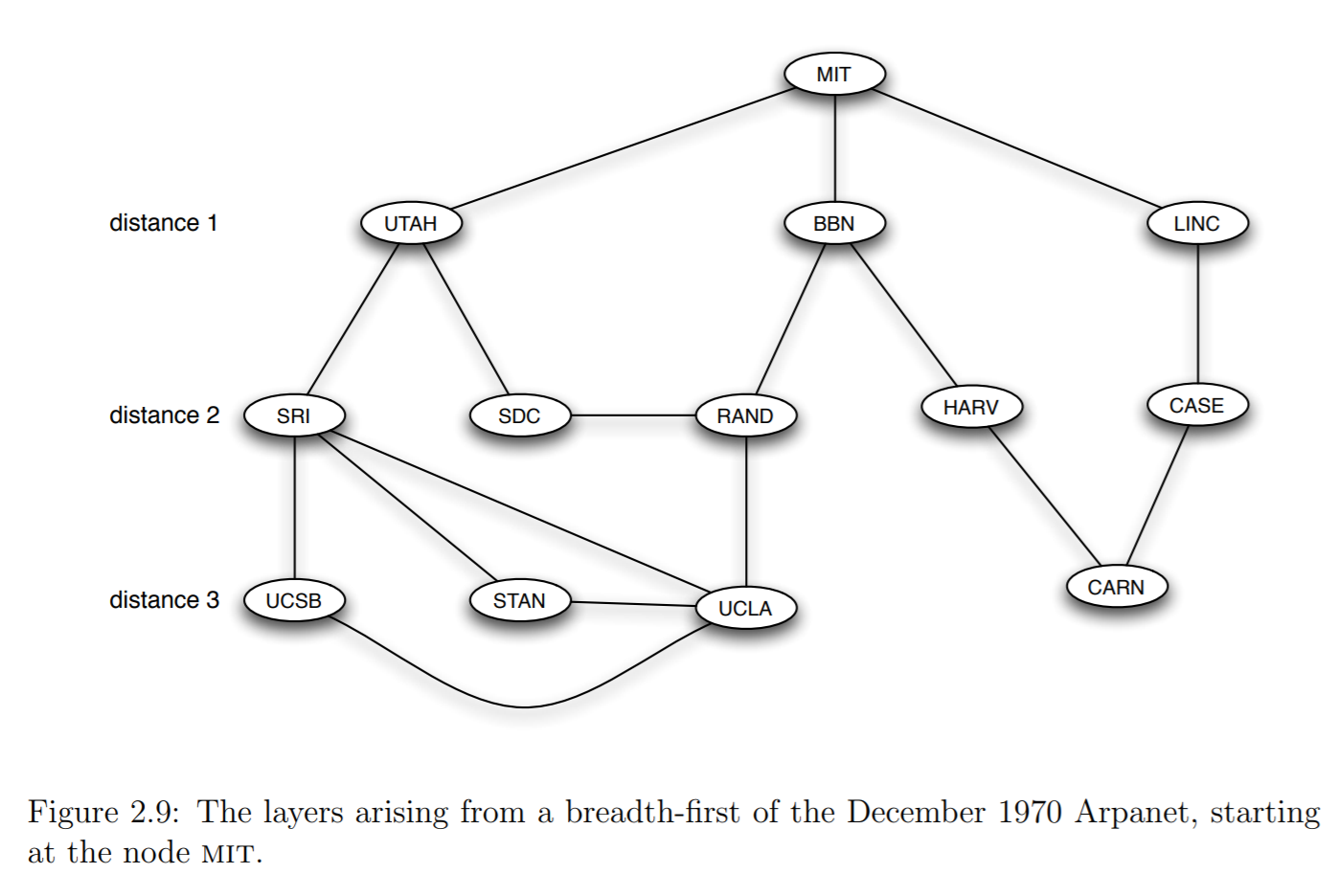
Exercise (difficult!): BFS¶
- Now it is time for you to try this out!
- Our goal is to use breadth-first search to compute the distance betwen a given node and all other nodes
- The return value you end up with should be an
Vector{Vector{Int}}, where elementiof this vector contains all node labels at distanceifrom the starting node - Fill in the logic for the
breadth_first_distancesfunction below
function breadth_first_distances(g, start::Int)
out = Vector{Int}[]
# use push!(out, new_nodes) to add to out
distance = 0
# TODO: your code here...
# return out
out
endbreadth_first_distances (generic function with 1 method)# Test code
function test_bfd_methods(val, want)
if length(val) == 0
error("Make sure to `push!` on to `out` in your function")
elseif length(val) != maximum(gdistances(arpa, node_ints["HARV"]))
error("`out` has incorrect number of elements")
elseif length.(val) != length.(want)
error("Right number of elements, but not right number in each subvector")
elseif all(map(x12 -> all(sort(x12[1]) .== sort(x12[2])), zip(val, want)))
println("correct!")
end
end
function run_tests()
val = breadth_first_distances(arpa, node_ints["HARV"])
want = [[9, 12], [7, 8, 11], [3, 6, 5, 10], [1, 2, 4]]
test_bfd_methods(val, want)
endrun_tests (generic function with 1 method)# uncomment the code below and run when you are ready to test your code
# run_tests()BFS with Graphs¶
- The Graphs library contains routines implementing breadth-first search
- The main function is called
bfs_tree
bfs_carn = bfs_tree(arpa, node_ints["CARN"]){13, 12} directed simple Int64 graph- Notice that the printout says we have a graph with 13 nodes, 12 edges and it is a directed graph
- Thus far, all graphs we have considered have been undirected
- We have only been concerned about if a connection (edge) exists between nodes
- A directed graph extends the notion of connecting nodes with a direction
- We can now say that things flow across edges from one node to another -- always in the same direction
- Why would the breadth-first search routine return a directed graph instead of the undirected type we started with?
- Let’s visualize it and see if we can understand why
gplot(bfs_carn, nodelabel=first.(nodes))Loading...
- Notice that arrows only flow out of
CARN - They also always flow away from
CARN - The use of directed edges allows Graphs to represent the shortest path from CARN to any other node
- For example
STAN:CARN -> HARV -> BBN -> RAND -> UCLA -> STAN
- For example
Exercise: Explore DiGraph¶
- The
bfs_carnobject has type $(Markdown.Code(string(typeof(bfs_carn)))) - Let’s view the names of its properties (properties)
propertynames(bfs_carn)(:ne, :fadjlist, :badjlist)bfs_carn.fadjlist13-element Vector{Vector{Int64}}:
[]
[]
[1, 2, 4]
[]
[]
[]
[3, 6]
[5]
[7, 8]
[]
[10]
[11, 13]
[9]- The
fadjlist(forward adjacency list) property is aVector{Vector{Int64}} fadjlisthas one element per node (call index into outer Vectorifor nodei)- Each element is itself a vector containing node indices for all nodes
jfor which there is an edge flowing fromitoj - Below we have set up a new method (see below) for the
breadth_first_distancesfunction that takes aDiGraphas an argument - Your task is to implement the the method so that it has the same return value as the previous method from above
function breadth_first_distances(g::SimpleDiGraph, start::Int)
out = Vector{Int}[]
# use push!(out, new_nodes) to add to out
distance = 0
# TODO: your code here...
# return out
out
endbreadth_first_distances (generic function with 2 methods)# test code
function test_digraph_ex()
val = breadth_first_distances(
bfs_tree(arpa, node_ints["HARV"]),
node_ints["HARV"]
)
want = [[9, 12], [7, 8, 11], [3, 6, 5, 10], [1, 2, 4]]
test_bfd_methods(val, want)
endtest_digraph_ex (generic function with 1 method)# uncomment the code below when you are ready to test your code!
# test_digraph_ex()Components¶
- A component of a graph is a self-contained subset of the nodes
- More precisely, a set of nodes is a component if
- Every node in the subset has a path to every other node in the subset
- The subset is not part of a larger set with property (1)
- Example:
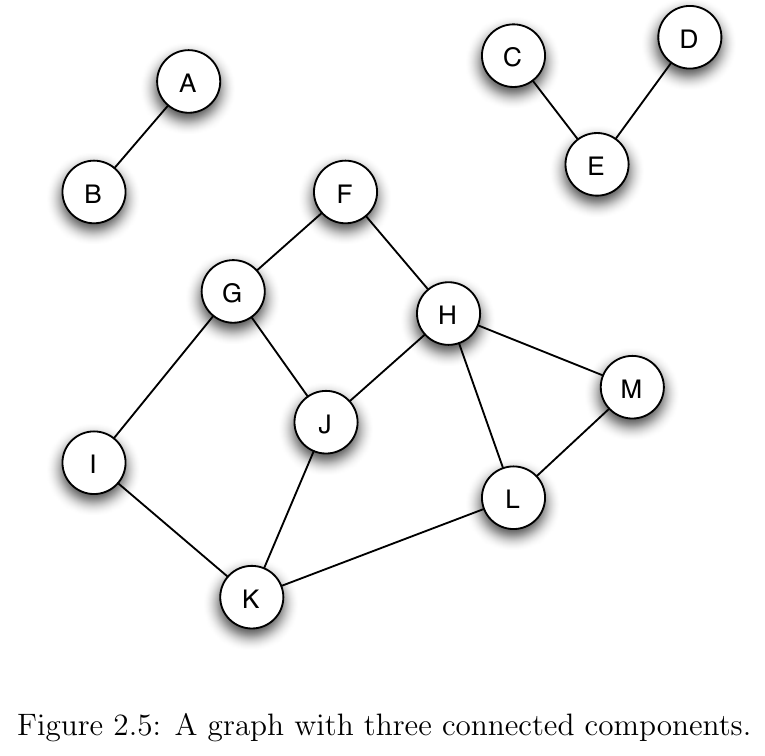
Finding Components¶
- Graphs.jl provides functions to identify and work with components
connected_components(g)returns a vector of vectors, where each inner vector contains the nodes in one component- Let’s create a graph with multiple components and explore it
# Create a graph with three components
multi_component = SimpleGraph(10)
# Component 1: nodes 1, 2, 3
add_edge!(multi_component, 1, 2)
add_edge!(multi_component, 2, 3)
add_edge!(multi_component, 3, 1)
# Component 2: nodes 4, 5, 6, 7
add_edge!(multi_component, 4, 5)
add_edge!(multi_component, 5, 6)
add_edge!(multi_component, 6, 7)
add_edge!(multi_component, 7, 4)
# Component 3: nodes 8, 9
add_edge!(multi_component, 8, 9)
# Node 10 is isolated (its own component)truecomponents = connected_components(multi_component)4-element Vector{Vector{Int64}}:
[1, 2, 3]
[4, 5, 6, 7]
[8, 9]
[10]println("Number of components: ", length(components))
for (i, comp) in enumerate(components)
println("Component $i has $(length(comp)) nodes: $comp")
endNumber of components: 4
Component 1 has 3 nodes: [1, 2, 3]
Component 2 has 4 nodes: [4, 5, 6, 7]
Component 3 has 2 nodes: [8, 9]
Component 4 has 1 nodes: [10]
gplot(multi_component, nodelabel=1:10)Loading...
Why Components Matter¶
- Components help us understand the structure of social networks
- Each component represents a group that can communicate internally but not with other groups
- In social network analysis, components reveal:
- Information silos: Ideas spread within components but not between them
- Echo chambers: Opinions reinforce within isolated groups
- Community detection: Natural groupings in social structures
- Influence boundaries: Limits of where influence can spread
Real-World Implications¶
- Disease spread: Each component represents a boundary for transmission
- COVID-19 travel restrictions aimed to keep countries as separate components
- Contact tracing identifies components to contain outbreaks
- Marketing: Products spread through word-of-mouth within components
- Need to seed each component separately for full market penetration
- Viral marketing campaigns can fail if they don’t bridge components
- Political polarization: Separate components in social media
- Different groups may never see opposing viewpoints
- Bridging nodes between components become crucial for dialogue
- Infrastructure resilience: Power grids, internet backbone
- Multiple components mean system fragmentation after failures
- Redundant connections prevent network from splitting into components
Giant Component¶
- Many real-world networks have a giant component containing most nodes
- Plus several small components or isolated nodes
- Let’s check if ARPANET has this structure
arpa_components = connected_components(arpa)
println("ARPANET has $(length(arpa_components)) component(s)")
println("Component sizes: ", length.(arpa_components))ARPANET has 1 component(s)
Component sizes: [13]
- ARPANET is fully connected (one component) by design
- This ensures any site can communicate with any other site
- Network resilience was a key design goal for the early internet
Real-World Network Examples¶
- Let’s analyze some actual network datasets to see these concepts in practice
- We’ll look at:
- Zachary’s Karate Club: A classic social network dataset
- Facebook ego networks: Social connections from real Facebook data
- Email networks: Communication patterns in organizations
Zachary’s Karate Club¶
- Famous dataset from sociology (Zachary, 1977)
- Documents friendships in a karate club that split into two factions
- 34 members (nodes), 78 friendships (edges)
- Perfect for studying community structure and components
# Graphs.jl includes several famous networks as built-in datasets!
karate = smallgraph(:karate){34, 78} undirected simple Int64 graph# Analyze the karate club network
using Statistics # for `mean` function
println("Karate Club Network:")
println(" Nodes: ", nv(karate))
println(" Edges: ", ne(karate))
println(" Average degree: ", round(mean(degree(karate)), digits=2))
println(" Is connected: ", is_connected(karate))Karate Club Network:
Nodes: 34
Edges: 78
Average degree: 4.59
Is connected: true
# Look at degree distribution
degrees = degree(karate)
println("\nDegree distribution:")
println(" Min degree: ", minimum(degrees))
println(" Max degree: ", maximum(degrees))
println(" Node with most connections: ", argmax(degrees))
Degree distribution:
Min degree: 1
Max degree: 17
Node with most connections: 34
# Visualize the karate club network
gplot(karate, nodelabel=1:nv(karate), nodesize=0.1)Loading...
Exercise: Analyzing the Petersen Graph¶
- The Petersen graph is a famous graph in graph theory
- It appears in many theoretical results and counterexamples
- Your task: analyze this network’s structure
# Load the Petersen graph
petersen = smallgraph(:petersen)
# TODO: Calculate the following properties
num_nodes = missing # Total number of nodes
num_edges = missing # Total number of edges
avg_degree = missing # Average degree (should be the same for all nodes!)
is_it_connected = missing # Is the graph connected?
is_it_tree = missing # Is it a tree?
num_components = missing # How many components?
# Print your results
println("Petersen Graph Analysis:")
println(" Number of nodes: ", num_nodes)
println(" Number of edges: ", num_edges)
println(" Average degree: ", avg_degree)
println(" Is connected: ", is_it_connected)
println(" Is it a tree: ", is_it_tree)
println(" Number of components: ", num_components)Petersen Graph Analysis:
Number of nodes: missing
Number of edges: missing
Average degree: missing
Is connected: missing
Is it a tree: missing
Number of components: missing
Other Built-in Networks¶
- Graphs.jl includes many classic networks for teaching and research
- Some interesting examples you can explore:
# Examples of other built-in graphs
graphs_to_explore = [
:house, # The "house" graph (5 nodes)
:bull, # The bull graph
:cubical, # 3-dimensional cube
:diamond, # Diamond graph
:dodecahedral, # Dodecahedron graph (20 nodes)
:heawood, # Heawood graph (14 nodes)
:moebiuskantor,# Möbius-Kantor graph
:octahedral, # Octahedron graph
:pappus, # Pappus graph
:tutte, # Tutte graph (46 nodes)
]
# Try loading and analyzing one!
# Example: g = smallgraph(:house)10-element Vector{Symbol}:
:house
:bull
:cubical
:diamond
:dodecahedral
:heawood
:moebiuskantor
:octahedral
:pappus
:tutte